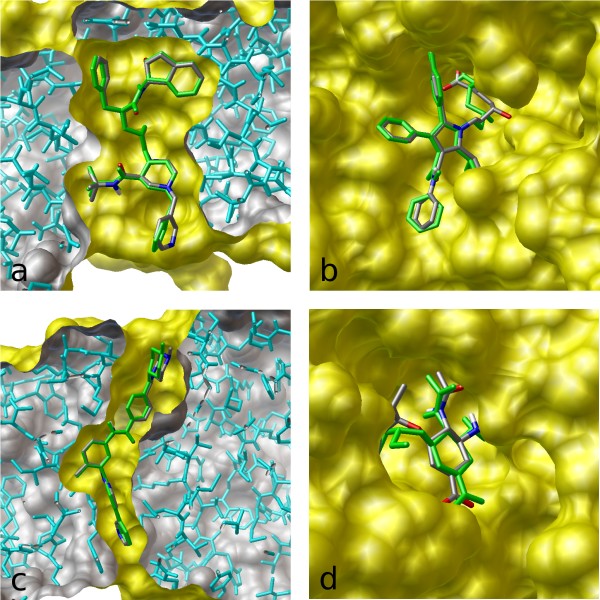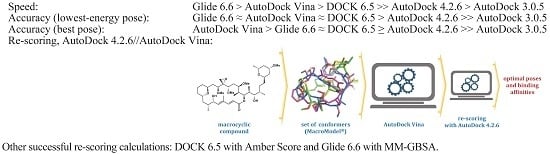
Molecules | Free Full-Text | The Performance of Several Docking Programs at Reproducing Protein–Macrolide-Like Crystal Structures

An accurate and universal protein-small molecule batch docking solution using Autodock Vina - ScienceDirect

Docking simulations identify potential compound binding sites in CT and... | Download Scientific Diagram

AutoDock Vina 1.1.2 – Molecular Docking and Virtual Screening Program – My Biosoftware – Bioinformatics Softwares Blog
Frontiers | ER/AR Multi-Conformational Docking Server: A Tool for Discovering and Studying Estrogen and Androgen Receptor Modulators
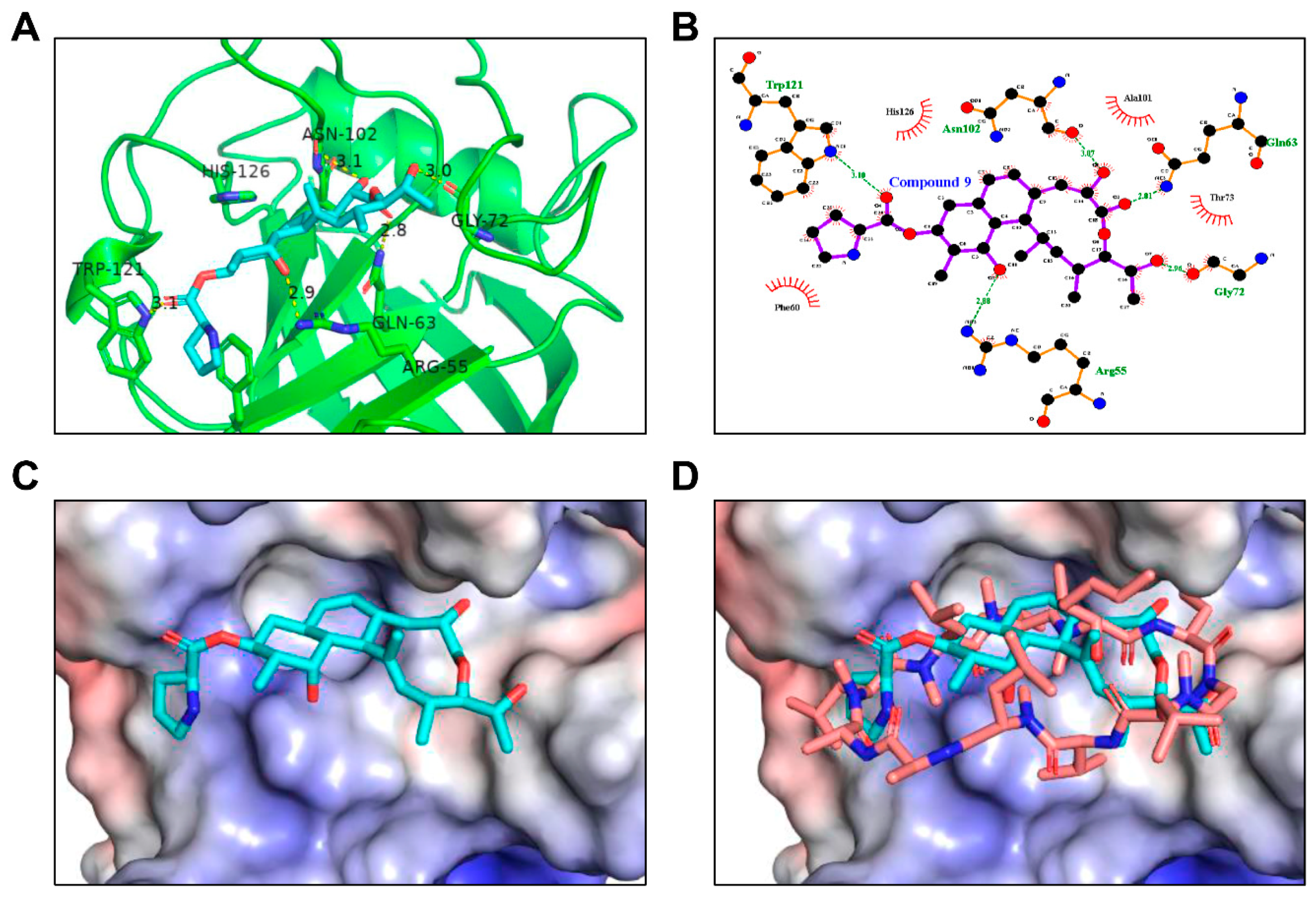
Targeting SARS-CoV-2 main protease by teicoplanin: a mechanistic insight by in silico studies Graphical Abstract
![PDF] Comparing AutoDock and Vina in Ligand/Decoy Discrimination for Virtual Screening | Semantic Scholar PDF] Comparing AutoDock and Vina in Ligand/Decoy Discrimination for Virtual Screening | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/d9c640af35595730416a5138c4feaa1af9bb7e03/9-Table2-1.png)
PDF] Comparing AutoDock and Vina in Ligand/Decoy Discrimination for Virtual Screening | Semantic Scholar

The interaction between JS-K and TAGLN were displayed using Autodock.... | Download Scientific Diagram

PDF) Performance Analysis of Embarassingly Parallel Application on Cluster Computer Environment: A Case Study of Virtual Screening with Autodock Vina 1.1 on Hastinapura Cluster | Muhammad Hilman - Academia.edu

Speed vs Accuracy: Effect on Ligand Pose Accuracy of Varying Box Size and Exhaustiveness in AutoDock Vina - Agarwal - 2023 - Molecular Informatics - Wiley Online Library

PLHINT: A knowledge-driven computational approach based on the intermolecular H bond interactions at the protein-ligand interface from docking solutions - ScienceDirect

AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings | Journal of Chemical Information and Modeling